Presence of unculturable bacteria in the permeate of microfiltration membranes with various pore-sizes in wastewater treatment
Abstract; Shuai Zhou, Taro Urase and Saki Goto. Journal of Water Process Engineering, 76, 108182. (2025). (DOI)
Constituents of Coliform Species Contained in the Permeate of Microfiltration Membranes in Wastewater Treatment
Abstract; Shuai Zhou, Taro Urase and Saki Goto. Water, 16(9), 1269. (2024). (DOI)
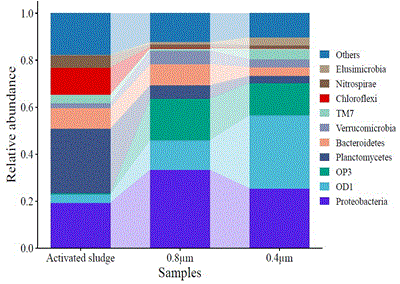
Bacterial Species Identified in the Filtrate of Microfiltration Membranes in the Separation of Activated Sludge
Abstract; Shuai ZHOU, Manae NINOSEKI, Asahi KUSABA, Kazuma NAKAGAWA, Taro URASE. Journal of Water and Environment Technology, Vol.19, No.6, Pages 294-301. (2021). (DOI)
Retention of Bacteria and DNA by Microfiltration Membranes in Wastewater Reclamation
Abstract; Hirofumi Tsutsui, Taro Urase. Journal of Water and Environment Technology, Vol. 17, No. 3: 194-202. (2019).(DOI)
Effect of antimicrobials in feed wastewater on the performance of two-stage membrane bioreactor
Taro URASE, Hirofumi TSUTSUI, Takeshi INOU, Hao Yang CHEN. Journal of Japan Society on Water Environment, Vol.40, No.3, pp.107-114. (2017).
Removal of color from molasses wastewater using membrane bioreactor with acidic condition
Ahmad SHAHATA, Takumi OMATA, Taro URASE. Journal of Water and Environment Technology, Vol. 11, No.6, 539-546 (2013).
Treatment of Saline Wastewater by Thermophilic Membrane Bioreactor
Ahmad Shahata, Taro Urase. J. of Water and Environment Technology, Vol. 14, No. 2, pp. 76-81 (2016). (DOI)
Factors affecting removal of pharmaceutical substances and estrogens in membrane separation bioreactor
Taro Urase, Chie Kagawa and Tomoya Kikuta. Desalination, 178, 107-113.(2005)